Common Mechanism for DNA Synthesis Screening
Last updated: July 2025
The Common Mechanism provides free, open-source, globally-available tools that help providers of synthetic DNA and RNA screen orders efficiently, securely, and in compliance with global biosecurity standards. These tools include:
- commec, a software package designed as a global baseline for sequence screening
- practical customer screening resources
- support for other technical resources to strengthen synthesis screening
IBBIS’s tools are hosted in Switzerland, developed by an international consortium, and incorporate biosecurity standards from every continent. This page aims to answer some frequently asked questions about the Common Mechanism.
Other Resources
- Webpage. Our Work: Common Mechanism. Available on ibbis.bio.
- Code. commec: a free, open-source, globally available tool for DNA sequence screening. Available on Github.
- Paper. Overcoming Challenges to Developing a Common Global Baseline for Nucleic Acid Synthesis Screening. Nicole Wheeler, Sarah R. Carter, Tessa Alexanian, Christopher Isaac, Piers Millett, Jaime Yassif. Applied Biosafety, April 2024. Available online (archive</a >).
- Whitepaper. Verifying Legitimacy: Findings from the Customer Screening Working Group, 2020-2023. Tessa Alexanian, Sarah R. Carter. February 2024. Available online.
Frequently Asked Questions
GENERAL
The tools are designed to help providers screen orders efficiently, securely, and in compliance with global biosecurity standards. These tools include a software package designed as a global baseline for sequence screening; practical resources for customer screening; and supports technical resources to strengthen synthesis screening. The Common Mechanism tools are hosted in Switzerland, developed by an international consortium, and incorporate biosecurity standards from every continent.
Synthesis screening remains voluntary, inconsistent, and globally fragmented; it is easy to find companies or intermediaries that do not screen, but new standards, tools and regulations have changed incentives around screening. Synthesis screening has long been necessary to comply with export control regulations in over 50 countries, bur is now recommended by national-level guidance in multiple countries, leading AI developers, ISO standards, and the WHO.
IBBIS works to increase the share of synthesis orders for which sequences and customers are screened and support international standards that are inclusive and rigorous. Several IBBIS projects address this challenge, including our work on the Common Mechanism, Global DNA Synthesis Map, and Screening Standards.- Free, open-source software and databases. The commec software package, and its associated databases are fully public and open-source.
- No data transfer. Intellectual property embedded in customer sequences can make providers reluctant to outsource screening to third-party services. The commec software is designed to be installed locally by individual providers, lowering barriers to access while protecting customer data.
- Globally accessible, internationally designed. The software was designed by an international technical consortium and incorporates screening standards from every continent.
- Customer screening. We also offer methods for verifying customer legitimacy, developed with a focus on serving international customers.
- January 2020: the Common Mechanism was proposed by an international working group jointly convened by the Nuclear Threat Initiative (NTI) and the World Economic Forum (WEF), which also called for an international entity that will house the mechanism, promote its adoption, and work to establish global norms for nucleic acid synthesis screening.
- 2020-2023: advised an international technical consortium, the Common Mechanism software and databases were developed by a team led by Dr. Nicole Wheeler of the University of Birmingham, and including contributions from Brittany Rife Magalis of the University of Louisville and Jennifer Lu of the Center for Computational Biology at Johns Hopkins University.
- February 2024: IBBIS is launched, acting as a long-term home for the Technical Consortium and Common Mechanism, among other initiatives to advance biosecurity.
- May 2024: public launch of the first open-source version of the Common Mechanism software.
FUNCTIONALITY AND USAGE
- Biorisk search: Sensitively identify well-established sequences of concern, such as toxins and virulence factors. (This is done using a fast HMM-based search against sequence profiles curated from annotations of regulated pathogens and toxins.)
- Taxonomy Search: Query large protein and DNA databases to retrieve the organism the genome most closely matched to the query sequence, then cross-referencing that match with a variety of control lists, including those from India, China, and South Africa.
- Low-concern search: Clear earlier flags based on matches to common or conserved sequences. This database includes protein sequences found in thousands of bacterial species, RNA sequences that participate in processes essential for life, and sequences submitted to the iGEM parts registry with no associated safety flags.
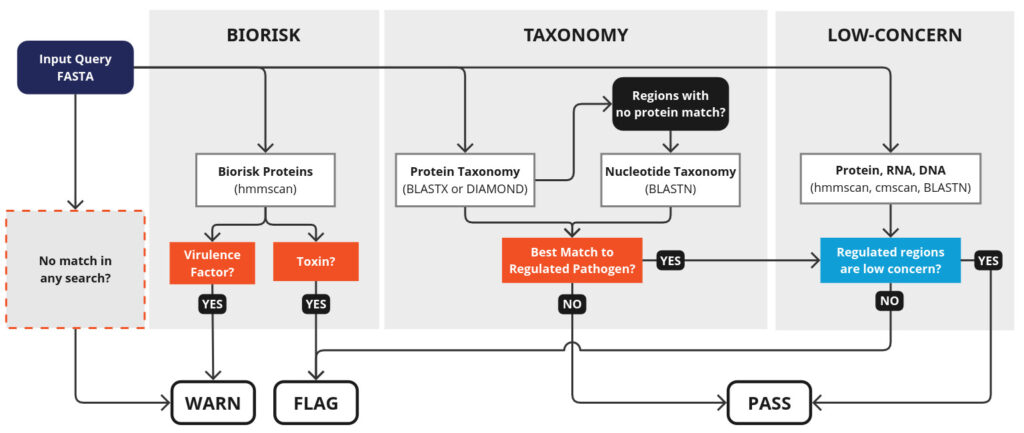
- Sequences below 50bp cannot be screened, and screening performance is best with sequences over 150bp.
- Protein sequences cannot be screened directly, though translations of the input DNA sequences are screened as part of the process.
- The system does not have any specialized functionality for screening oligo orders.
- Sequences containing ambiguity codons may not be scored correctly by every step in the pipeline.
GETTING STARTED
PRIVACY AND SECURITY
GUIDELINES AND COMPLIANCE
TROUBLESHOOTING
• Processing speed: When running comprehensive searches using NCBI databases, the software may require significant processing time depending on your computational resources.
• Database size requirements: The software depends on large reference databases (~600 GB for standard protein and nucleotide databases), which may present storage challenges for some users.
• Oligonucleotide handling: The system does not currently have specialized functionality for screening oligo orders.
• Scope: The Common Mechanism is focused solely on sequence screening and does not provide customer screening capabilities. For guidance on customer screening, please refer to our customer legitimacy verification guidelines.
IBBIS is constantly working on reducing these limitations, which should gradually disappear as new updates are released.ATTRIBUTION
April 2024. Overcoming Challenges to Developing a Common Global Baseline for Nucleic Acid Synthesis Screening.Nicole Wheeler, Sarah R. Carter, Tessa Alexanian, Christopher Isaac, Piers Millett, Jaime Yassif. Applied Biosafety. Available online (archive).
 or the horizontal wordmark (also available for light backgrounds):
or the horizontal wordmark (also available for light backgrounds):

